
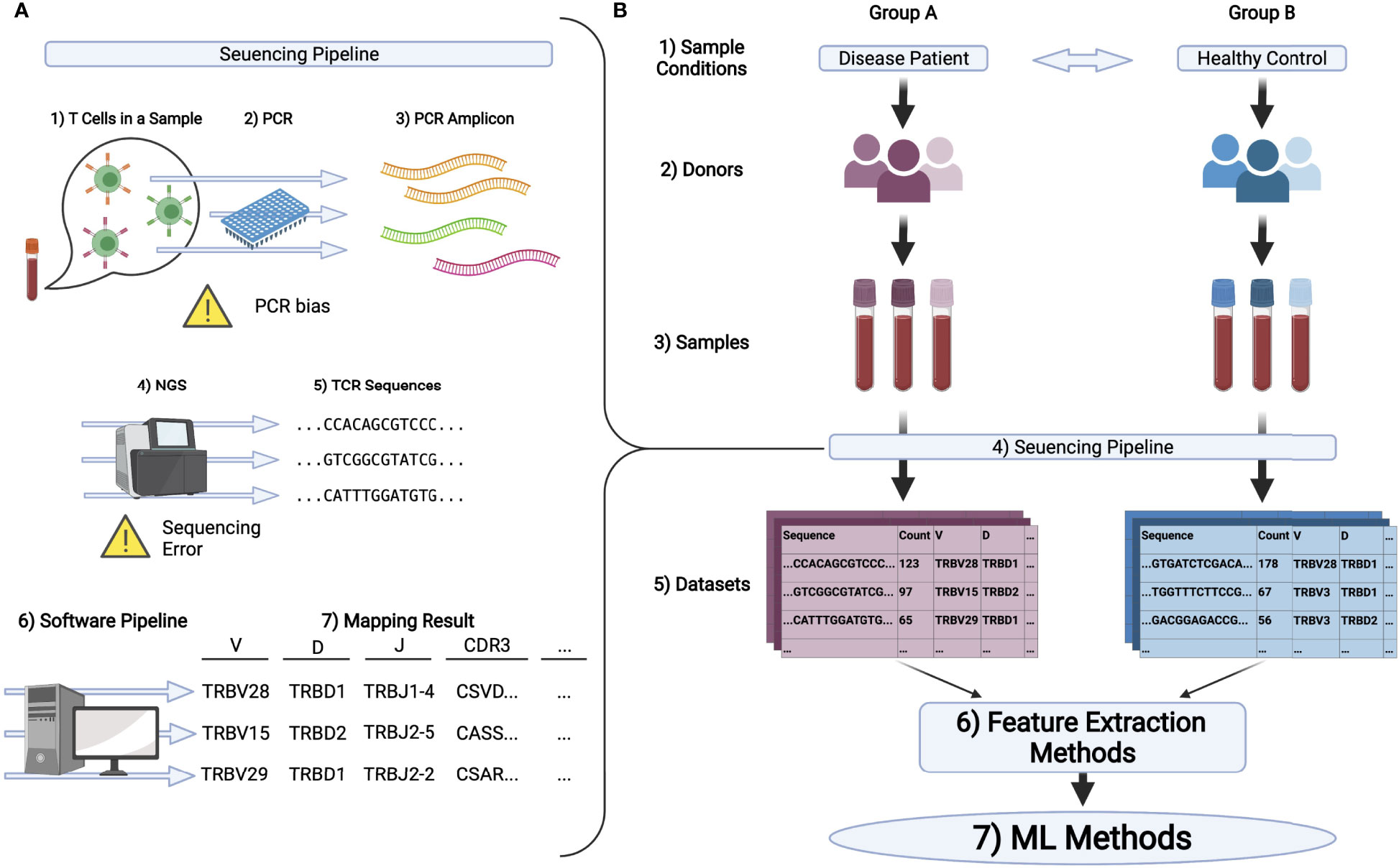
( d, e) Heatmap of top differentially expressed genes between T cell ( d) and B cell ( e) clones with either one cell or clonally expanded. ( c) CD4 and CD8a expression for those clones supported by a single cell or those expanded clones (more than 1 cell). Clonally expanded T and B cells correspond to those clones (identical, paired CDR3 amino acid sequence) containing two or more distinct cell barcodes. ( b) Cluster membership of clonally expanded T and B cells in V(D)J libraries. Clone is defined as identical, paired amino acid CDR3 sequences (CDR β3 + CDR α3 for T cells, CDRH3 + CDR元 for B cells) with exactly one heavy chain and one light chain. Expanded B and T cells were defined by those clones containing at least 2 distinct cell barcodes.
( a) Location and cluster membership of expanded B and T cells. (Online version in colour.)Ĭlonally expanded lymphocytes have distinct transcriptional profiles. ( f) Normalized gene expression of for B cells ITGAX, AID (AICDA), CD30 (TNFRSF8), T-bet (TBX21), CD138 (SDC1) and CD19. ( e) Normalized gene expression of for T cells for PD-1 (PDCD1), LAG3, CTLA4, TIM3 (HAVCR2), TBET (Tbx21), IFNG, Granzyme B (GZMB) and CD44. Cells are ordered by p-value after adjusting for multiple hypothesis testing. Intensity corresponds to normalized gene expression. Displayed cells were randomly sampled from within each cluster proportional to cluster's total number of cells. Clusters were calculated separately for B cells and T cells. ( c, d) Heatmap displaying differentially expressed genes between young and 18-month-old T and B cells, respectively. Clusters were determined using either only T cells or only B cells found in V(D)J libraries. ( b) The percentage of T (top) or B (bottom) cells found within each cluster for each of the experimental cohorts. Colour corresponds to transcriptional cluster specific to T or B cell analysis. ( a) Unsupervised clustering and uniform manifold approximation projection (UMAP) was performed separately on either B or T cells that were also detected in V(D)J libraries. Transcriptional signatures of clonally expanded lymphocytes. Both HC and LC sequences correspond to full-length V and J regions. ( g) The mean number of nucleotide substitutions per B cell clone detected on either the HC or light chain LC. ( f) The mean number of nucleotide somatic hypermutations (SHMs) per B cell clone in the full-length V and J regions across both heavy (HC) and light chain (LC). Variant is defined as unique, combined V H + V L amino acid sequence. ( e) The number of somatic variants per expanded B cell clone. Horizontal line within the box indicates the median and dot above. ( d) Tukey box-plot displaying the number of unique molecular identifiers (UMI) per those clones with reads mapping exclusively to the IgD isotype. ( c) Number of clones with exclusively IgD BCR transcripts. ( b) Isotype distribution for those expanded (greater than 1 cell barcode) B cell clones. For those clones with multiple cells, the majority isotype was selected.
( a) Percentage of unique clones with a given BCR. Our findings implicate that clonally related T and B cells in the CNS of elderly mice may contribute to neuroinflammation accompanying homeostatic ageing.īioinformatics central nervous system immune repertoires single cell.ī cells in the CNS are primarily of the IgM isotype with minor somatic hypermutation. Integrating gene expression information additionally revealed distinct transcriptional profiles of these clonally expanded lymphocytes. Furthermore, many of these B cells were of the IgM and IgD isotypes, and had low levels of somatic hypermutation. We observed the presence of clonally expanded B and T cells in the central nervous system of aged male mice. Here, we leveraged single-cell sequencing to simultaneously profile B cell receptor and T cell receptor repertoires and accompanying gene expression profiles in young and old mouse brains. Technical limitations, however, have prevented an integrative analysis of how lymphocyte immune receptor repertoires and their accompanying transcriptional states change with age in the central nervous system. Neuroinflammation plays a crucial role during ageing and various neurological conditions, including Alzheimer's disease, multiple sclerosis and infection.


 0 kommentar(er)
0 kommentar(er)
